




🔵 Projects We Lead
🟠 Projects We Co-Lead
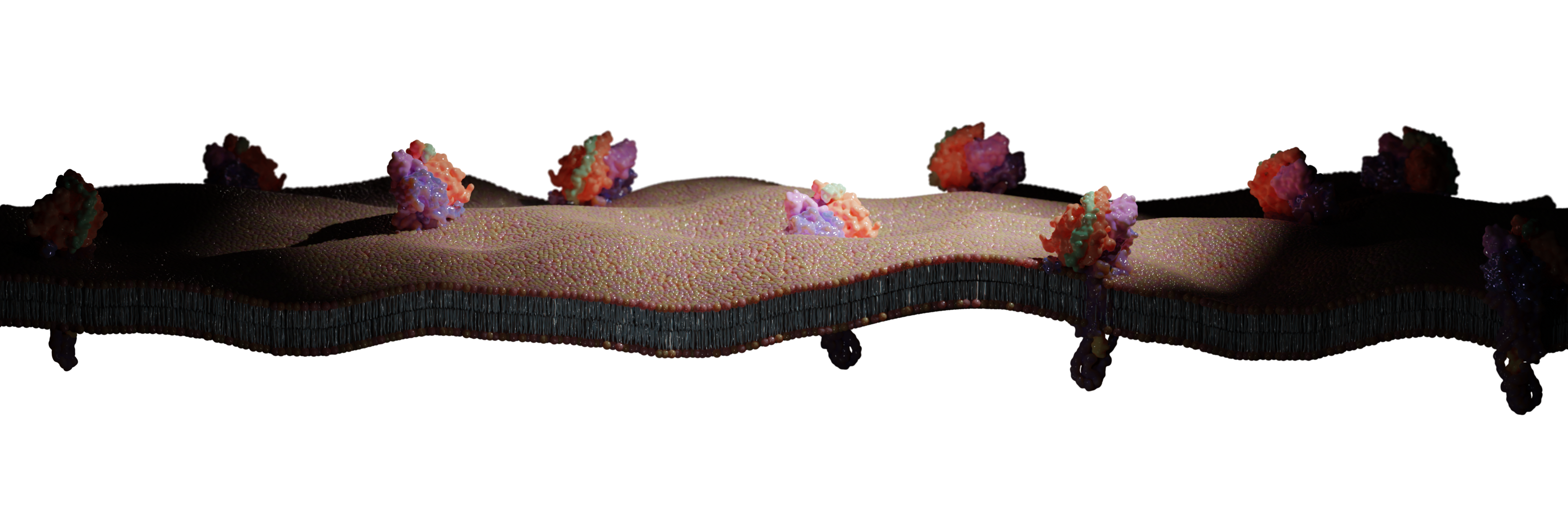
Establishing a Pan-European Network on Computational Redesign of Enzymes - COZYME
COST Action CA21162: Management Committee (2022)
TWIN2PIPSA - Twinning for Excellence in Biophysics of Protein Interactions and Self-Assembly
HORIZON-WIDERA-2021-ACCESS-03
Ruthenium-peptide conjugates: arrows for selectively targeting breast cancer
PTDC/QUI-QIN/0146/2020
Understanding and exploiting the impacts of low pH on micro-organisms - EuroMicropH
CMST COST Action CA18113 (2019)
New diagnostic and therapeutic tools against multidrug resistant tumors - Stratagem
CMST COST Action CM17104 (2018)
Uncovering blind spots in halogen bonding applications
PTDC/QUI-QFI/28455/2017
Multivalent Glycosystems for Nanoscience - MultiGlycoNano
CMST COST Action CM1102: Management Committee (2012)
Aumentando o realismo da modelação de membranas em métodos de dinâmica molecular a pH constante: inclusão de gradientes electroquímicos e titulação de lípidos
PTDC/QEQ-COM/1623/2012
Complexos de metais de transição usados como intercaladores de DNA: investigação teórica
Bilateral Program: PESSOA (2011)
Síntese, estudos computacionais e propriedades biológicas de compostos de ouro
Bilateral Program: FCT-CSIC (2010)
Understanding structure-activity relationships in peptide dendrimers using a molecular modelling approach
PTDC/QUI-QUI/100416/2008
Including protonation effects in the simulation of peptides and proteins in membrane environments
PTDC/BIA-PRO/104378/2008
Study of pH-dependent protein misfolding using state-of-the-art molecular modeling methods
PTDC/QUI-BIQ/105238/2008
© 2023 MMS@FCUL. All Rights Reserved. Crafted by João Vitorino